 Given a collection of “query” sequences read from an sample, what are an appropriate set of database sequences to use for a comparative analysis?
The traditional answer to this question has been “all of them that might be relevant”, which usually either means taking every best BLAST hit of the query sequences in some database, or the whole database itself.
As sequencing runs and databases both get bigger at a remarkable rate, this may no longer be a viable answer.
Given a collection of “query” sequences read from an sample, what are an appropriate set of database sequences to use for a comparative analysis?
The traditional answer to this question has been “all of them that might be relevant”, which usually either means taking every best BLAST hit of the query sequences in some database, or the whole database itself.
As sequencing runs and databases both get bigger at a remarkable rate, this may no longer be a viable answer.
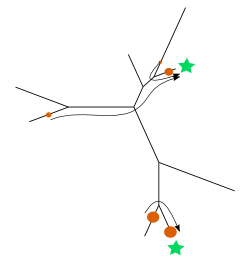
We decided to come up with a principled way of choosing an optimal set of k “reference” database sequences that are relevant for an analysis of the set of query sequences. The idea is very simple: build a tree on all of the potential sequences (or place the reads onto a big reference tree), and then choose the set of k sequences X that minimize the average distance from each query sequence to its closest reference sequence in the chosen set X. This ends up being a problem of optimal mass transport on trees (top image): imagine the tree being a road network, the query sequences being dirt piles on the road network, and the leaves of the tree being terminals. Now, given a k > 0, what is the selection of k terminals minimizing the total work (mass times distance) required to move the mass to the one of the k selected terminals (top image)?
This is related to the question of finding medoids and we can adapt a heuristic algorithm from that setting (called PAM) to solve the problem.
However, the PAM algorithm does not always find the optimal solution, motivating an exact algorithm.
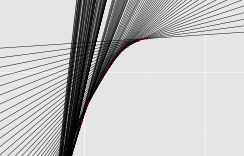
We have developed such an algorithm using ideas from convex geometry, in particular the double description algorithm to find lines that actually bound a polytope (lower image).
This project was fun fun fun, and has already been quite useful for our work in HIV superinfection, as well as for microbiome reference set creation.
The paper describing this method, as well as showing via simulation that this procedure avoids chimeric sequences, is submitted and up on arXiv.
All of these are coded up in the rppr binary of the pplacer suite.