 We were happy this week to see our new paper come out on bacterial communities in women with bacterial vaginosis, as surveyed through 16s 454 sequencing.
In contrast to some other previous studies, the explicit aim of this study was to study communities in women with BV.
We were happy this week to see our new paper come out on bacterial communities in women with bacterial vaginosis, as surveyed through 16s 454 sequencing.
In contrast to some other previous studies, the explicit aim of this study was to study communities in women with BV.
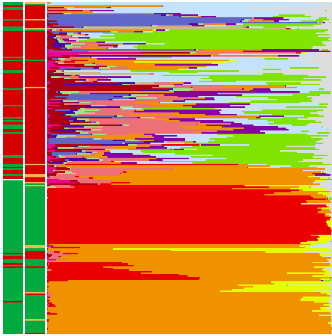
A lot of the story of the paper can be told in the figure here to the right. The two strips on the left show BV status by two different methods of evaluation, with red being positive and green being negative. The big bar plot is an abundance plot of the different species (represented by the different colors). Thus, as reported by a number of other groups, we see that BV is characterized by a highly diverse collection of taxa. This includes the BVAB’s, three species of Clostridium, which were discovered by the Fredricks lab.
This paper featured the use of the pplacer suite of tools in a variety of contexts. Of particular interest to our collaborators were species-level taxonomic classifications using phylogenetic placement of reads (paper in preparation). The paper also used our edge PCA and squash clustering methods to uncover interesting structure in the data. Time for us to get these methods published in their final versions!