 Our fourth paper coming from our collaboration with Lisa Jones-Engel and Maxine Linial has come out.
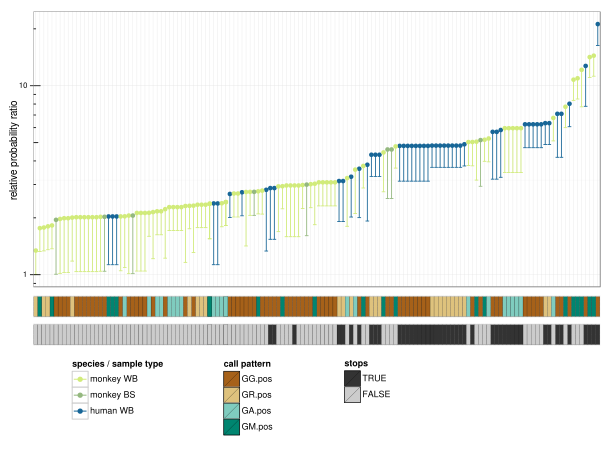
This paper shows that the human restriction factor APOBEC3G acts (perhaps along with other restriction factors) to protect us from extensive replication of SFV in vivo by inducing G to A mutations in SFV genomes.
We see extensive and strong hypermutation in human host SFV sequences that we don’t see in the monkey host SFV sequences, and we also see a different pattern of nucleotide context preference.
Among other things, this is the first (although indirect) evidence that SFV undergoes at least one round of replication in zoonotically infected humans.
Our fourth paper coming from our collaboration with Lisa Jones-Engel and Maxine Linial has come out.
This paper shows that the human restriction factor APOBEC3G acts (perhaps along with other restriction factors) to protect us from extensive replication of SFV in vivo by inducing G to A mutations in SFV genomes.
We see extensive and strong hypermutation in human host SFV sequences that we don’t see in the monkey host SFV sequences, and we also see a different pattern of nucleotide context preference.
Among other things, this is the first (although indirect) evidence that SFV undergoes at least one round of replication in zoonotically infected humans.
We have been working on this project for quite a while– for example, this picture shows the early days of using a print-out and ruler to identify suspicious G to A mutations. Since that time, we have gotten more sophisticated in our analysis. In fact, much of the paper is dedicated to our way of analyzing the 2x2 tables that come from evaluating a sequence for the presence of hypermutation. We were motivated to come up with a new way of doing so after reading about problems with applying the Fisher exact test when counts are small (which is when it gets applied). We heard about these issues from my neighbor Rob Bradley who in turn had heard about it from Yarden Katz.
In any case, we came up with a Bayesian approach whereby we estimate the ratio of the mutation rate in an APOBEC3-associated nucleotide context divided by the mutation rate outside of such a context. By directly estimating a posterior on this ratio, we get a better effect size estimate than the classical risk ratio (a.k.a. relative risk). We can use the same framework to estimate significance, which generates an appropriate tail distribution under the null. It also points the way towards generalization (stay tuned).