How frequently are genes transferred horizontally? A popular means of addressing this question involves building phylogenetic trees on many genes, and looking for genes that end up in surprising places. For example, if we have a lineage B that got a gene from lineage A, then a tree for that gene will have B’s version of that gene descending from an ancestor of A, which may be on the other side of the tree.
Using this approach requires that we have accurate trees for the genes. That means doing a good job with our modeling and inference, but it also means having data with plenty of the mutations which give signal for tree building. Unfortunately, sometimes we don’t have such rich data, but we’d still like to do such an analysis.
A naïve approach is just to run the sequences we have through maximum-likelihood tree estimation software and take the best tree for each gene individually, figuring that this is the best we can do with our incomplete data. However, noisy estimates with our sparse data will definitely bias our estimates of the impact of horizontal gene transfer (HGT) upwards. That is, if we get trees that are inaccurate in lots of random ways, it’s going to look like a lot of HGT.
We can do better by adding extra information into the inferential problem. For example, in this case we know that gene evolution is linked with species evolution. Thus in the absence of evidence to the contrary, it makes sense to assume that the gene tree follows a species tree. From a statistical perspective, this motivates a shrinkage estimator, which combines other information with the raw data in order to obtain an estimator with better properties than estimation using the data alone.
One way of doing this is to take a Bayesian approach to the full estimation problem, which might involve priors on gene trees that pull them towards the species tree using a generative model; this approach has been elegantly implemented in phylogenetics by programs such as PHYLDOG, SPIMAP and *BEAST. These programs are principled yet somewhat computationally expensive.
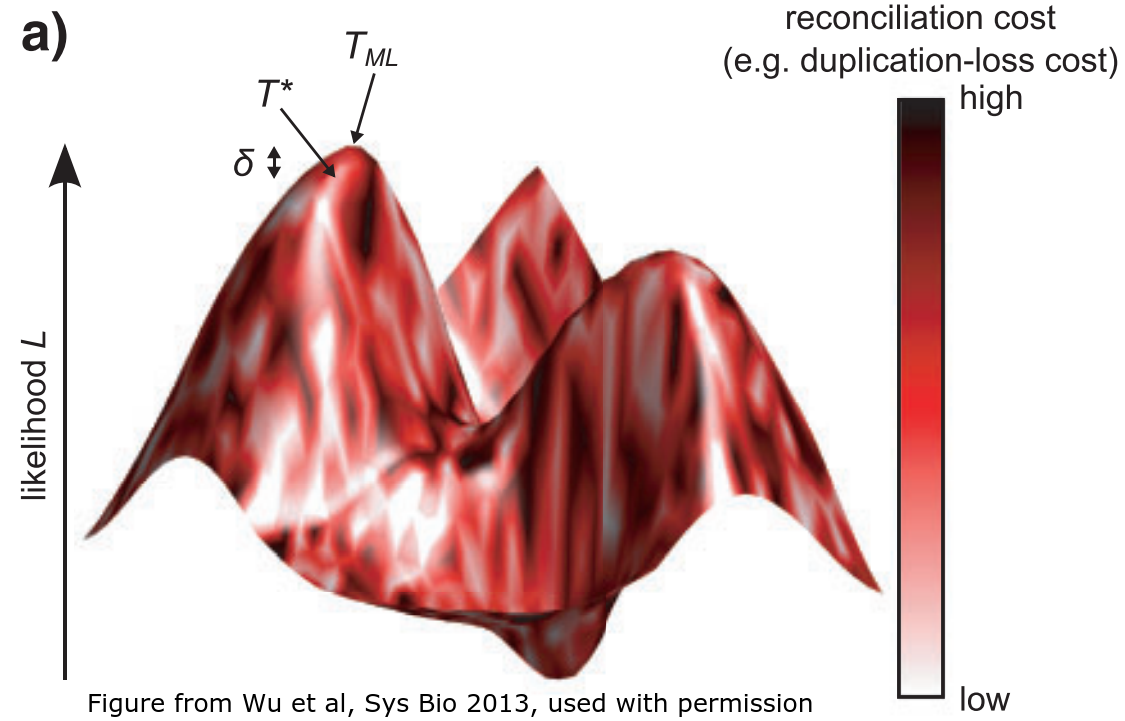
Another direction involves taking some sort of distance between the gene and species tree, and working to trade off a good value of the phylogenetic likelihood versus a good (small) value for this distance. This distance-based approach works surprisingly well! A 2013 paper by Wu et al proposed a method called TreeFix, which they showed performed almost as well as full SPIMAP inference, even in simulations under the SPIMAP generative model! The cartoon above is from their paper, and illustrates that it makes sense to trade off some likelihood (height) for a lower reconciliation cost (lighter color).
This definitely got my attention, and made me wonder if one could develop relevant theory, as the theoretical justification for such an approach doesn’t follow “automatically” like it does for a procedure doing inference under a full probabilistic model. Justification also doesn’t follow from the usual statistical theory for regularized estimators, because trees aren’t your typical statistical objects. Then, one day Vu’s high-school and college friend Lam Si Tung Ho was visiting, and I suggested this problem to them. They crushed it. What resulted went far beyond what I originally imagined: a manuscript that not only provides a solid theoretical basis for penalized likelihood approaches in phylogenetics, but also develops many useful techniques for theoretical phylogenetics. We have just put the manuscript up on arXiv, which develops a likelihood estimator which is regularized in terms of the Billera-Holmes-Vogtman (BHV) distance to a species tree.
First, the main results. The regularized estimator is “adaptive fast converging,” meaning that it can correctly reconstruct all edges of length greater than any given threshold from gene sequences of polynomial length. Perhaps more remarkable, though, is that Vu and Lam have explicit bounds of the convergence of the estimation simultaneously in terms of both branch length and topology (via the BHV distance). This goes beyond the standard theoretical phylogenetics framework of “did we get the right topology or not”. Surprisingly, the theory and bounds all work even if the species tree estimate is distant from the true gene tree, though of course one gets tighter bounds if it is close to the true gene tree.
Second, the new theoretical tools.
- a uniform (i.e. not depending on the tree) bound on the deviation of the likelihood of a collection of sequences generated from a model to their expected value
- an upper bound on the BHV distance between two trees based on the Kullback-Leibler divergence between their expected per-site likelihood functions
- analysis of the asymptotics of the regularization term close and far from the species tree.
Of course, I don’t think that biologists will plug in their desired error into our bounds and just sequence an amount of DNA required to achieve that level of error. That’s absurd. What I hope is that this paper will add a theoretical aspect to the body of evidence that regularization is a principled method for phylogenetic estimation, and help convince phylogenetic practitioners that raw phylogenetic estimates are inherently limited. We have all sorts of additional information these days that we can use for phylogenetic inference– let’s use it! Just ask your local statistician: that community has been impressed by the surprising effectiveness of regularization since the 50’s, and regularization in various forms has become a mainstay of modern statistical inference.