Immune receptor sequencing is stochastic through and through. We have cells with random V(D)J rearrangements that are stimulated through some random process of exposures, which lead to some random amount of expansion, and in the B cell case there is some random process of mutation and selection. So why don’t we use methods incorporating that uncertainty into our analysis?
We’ve tried to do this in our work, and have made some progress, but there is so much left to be done. When Sarah Cobey and Patrick Wilson kindly invited me to contribute to their special issue of Immunological Reviews, I knew I wanted to step back and ask:
If computation was no barrier, how would we design an analysis framework that integrated out uncertainty in unknown quantities and took advantage of the hierarchical structure inherent in immune receptor data?
I teamed up with Branden Olson, a Statistics PhD student in the lab, and went to work. It was a fun exercise to think through all of the steps of immune repertoire development and ask: what is the most realistic model under which inference should be possible, and what is the most realistic model for which we can perform simulation? This was more effort than anticipated, but 230 references later the final version is now up on arXiv and accessible for free (though I understand if you want to wait a few months to pay $38 and get it from the journal website).
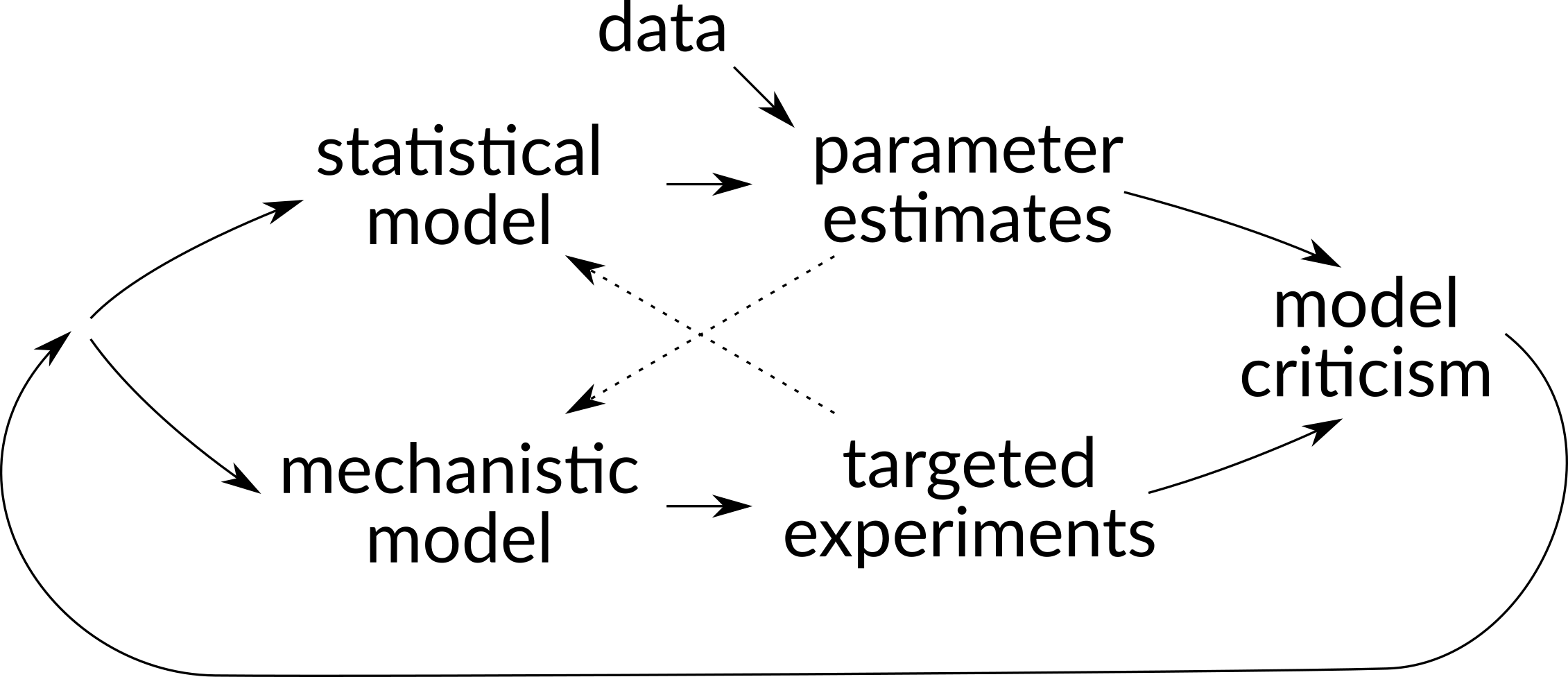
In addition to dreaming research directions, I wanted to explain to my immunologist pals why I think probabilistic analysis methods are crucial, and describe the basics of Bayesian analysis via simple metaphors. Ideally this will lead to a little more crosstalk between communities. Traditionally, statisticians and lab biologists have been on independent tracks (see image above) even though they investigate the same underlying phenomena. I hope that in the future we can unify these tracks by developing statistical models based on mechanism and design experiments based on statistical inferences.
I also hope that this serves as an invitation to the computational statistics community. As we say at the end: “The computational statistician interested in immune receptor modeling is blessed with a complex biological system to analyze, intractable computational problems heaped on top of one another, and an ever-expanding collection of data sets generated from various in-vivo and in-vitro perturbations.”
Come play!